SFtools: Space Filling Based Tools for Data Mining
SFtools is an R package available on CRAN and GitHub. It contains space filling based tools for machine learning and data mining. Basically, the main function proposes a new filter algorithm for the unsupervised feature selection problems. This algorithm is based on a space filling criterion called the coverage measure. Further updates will be added later.
This tutorial explains the use of this library with some simulated datasets and some real case studies. To download the library or get the code please visit the GitHub repository: https://github.com/mlaib/SFtools.
Version
1.0.0
Authors
Mohamed Laib and Mikhail Kanevski
Maintainer
Mohamed Laib [laib.med (at) gmail.com]
URL
https://cran.r-project.org/package=SFtools
License
GPL-3
Note
To cite this work, please use one of the following:
-
M. Laib, M. Kanevski, A novel filter algorithm for unsupervised feature selection based on a space filling measure. Proceedings of the 26rd European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning (ESANN), pp. 485-490, Bruges (Belgium), 2018.
-
M. Laib and M. Kanevski, A new algorithm for redundancy minimisation in geo-environmental data, 2019. Computers & Geosciences, 133 104328.
Use it
# Install it from CRAN:
install.packages("SFtools")
# Or from GitHub:
# First, you need to install: Biobase
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("Biobase")
devtools::install_github("mlaib/SFtools")
library(SFtools)
Numerical experiments
Application to simulated dataset
Butterfly dataset
library(IDmining)
dat <- Butterfly(1000)
dat<-dat[,-9]
Results<- UfsCov(dat)
## ===========================================================================
cou<-colnames(dat)
nom<-cou[Results[[2]]]
par(mfrow=c(1,1), mar=c(5,5,2,2))
names(Results[[1]])<-cou[Results[[2]]]
plot(Results[[1]] ,pch=16,cex=1,col="blue", axes = FALSE,
xlab = "Added Features", ylab = "Coverage measure")
lines(Results[[1]] ,cex=2,col="blue",lwd=2)
grid(lwd=1.5,col="gray" )
axis(2)
axis(1,1:length(nom),nom)
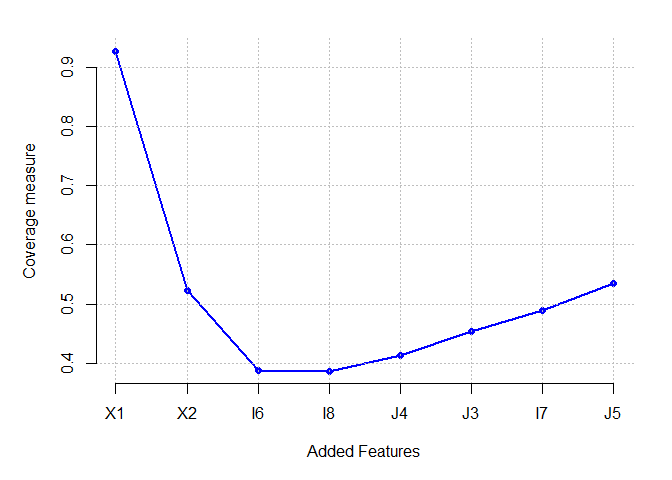
which.min(Results[[1]])
## I6
## 3
Real case studies
Parkinson dataset
raw_data <- read.csv("Parkinson.csv")
data<-raw_data[,1:22]
data <- unique(data)
names(data)<-c("V1","V2","V3","V4","V5","V6","V7","V8","V9","V10","V11","V12",
"V13","V14","V15","V16","V17","V18","V19","V20","V21","V22")
res<-UfsCov(data)
## ===========================================================================
par(mfrow=c(1,1), mar=c(5,4,1,1))
plot(res[[1]] ,pch=16,cex=1,col="blue", xlab = "Added Features",
ylab = "Coverage measure", axes=F)
lines(res[[1]] ,cex=2,col="blue")
axis(1)
axis(2)
abline(h=min(na.omit(res[[1]])))
grid(lwd=1.5,col="gray" )
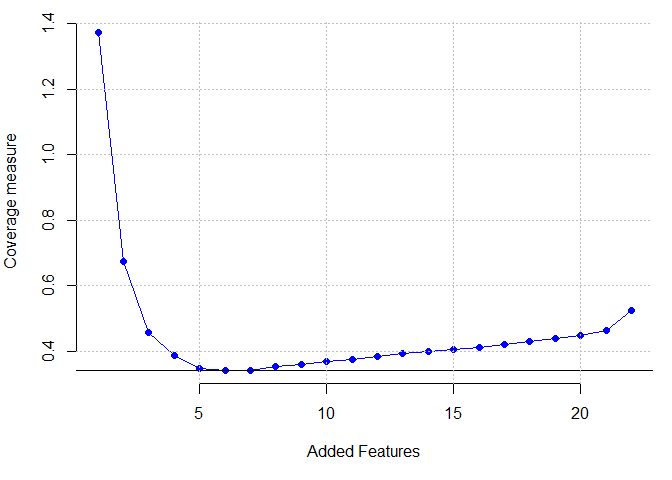
which.min(res[[1]])
## [1] 7
Pageblock dataset
raw_data <- read.table(file="PageBlocks.csv",sep=",")
colnames(raw_data)<-c("height","lenght","area","eccen","p_black","p_and","mean_tr",
"blackpix","blackand","wb_trans","target")
data<-raw_data[,1:10]
data <- unique(data)
res<-UfsCov(data)
## ===========================================================================
par(mfrow=c(1,1), mar=c(5,4,1,1))
plot(res[[1]] ,pch=16,cex=1,col="blue", xlab = "Added Features", ylab = "Covering measure", axes = FALSE)
axis(1)
axis(2)
lines(res[[1]] ,cex=2,col="blue", lwd=2)
abline(h=min(na.omit(res[[1]])))
grid(lwd=1.5,col="gray" )
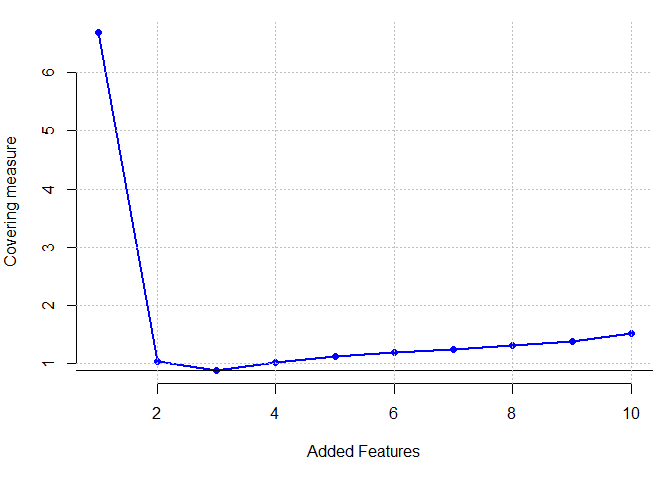
which.min(res[[1]])
## [1] 3
Ionsphere dataset
raw_data <- read.table(file="Ionosphere.csv",sep=",")
data<-raw_data[,1:34]
data <- unique(data)
data<-data[,-2]
res<-UfsCov(data)
## ===========================================================================
par(mfrow=c(1,1), mar=c(5,5,2,2))
plot(res[[1]] ,pch=16,cex=1,col="blue", xlab = "Added Features", ylab = "Covering measure", axes = FALSE)
axis(1)
axis(2)
lines(res[[1]] ,cex=2,col="blue", lwd=2)
abline(h=min(na.omit(res[[1]])))
grid(lwd=1.5,col="gray" )
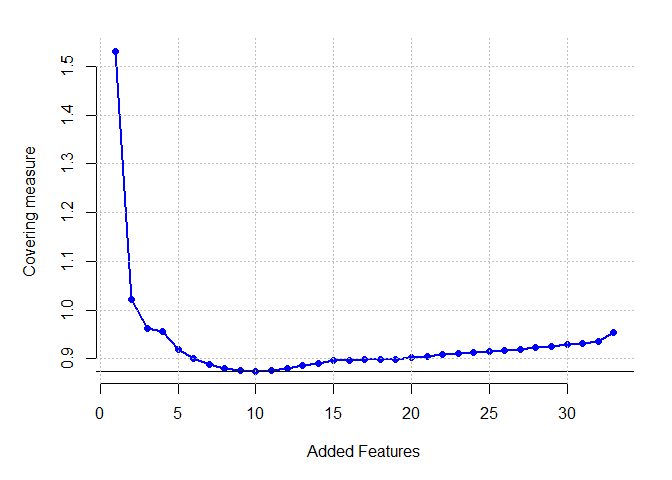
which.min(res[[1]])
## [1] 10